ELIXIR Norway
ELIXIR Norway is a national node of ELIXIR, the pan-European infrastructure for biological information, supporting life science research and its translation to medicine, environment, the bioindustries and society.
ELIXIR Norway is funded by the Research Council of Norway, and builds upon the Norwegian Bioinformatics Platform, which has provided support and tools for Norwegian life science research for nearly two decades.
Helpdesk
Short- and long-term support with bioinformatics analyses, programming and data management tasks. Contact the helpdesk.
Services
Analysis and management of life science data within marine, health, genomics, proteomics, structural analysis and more.
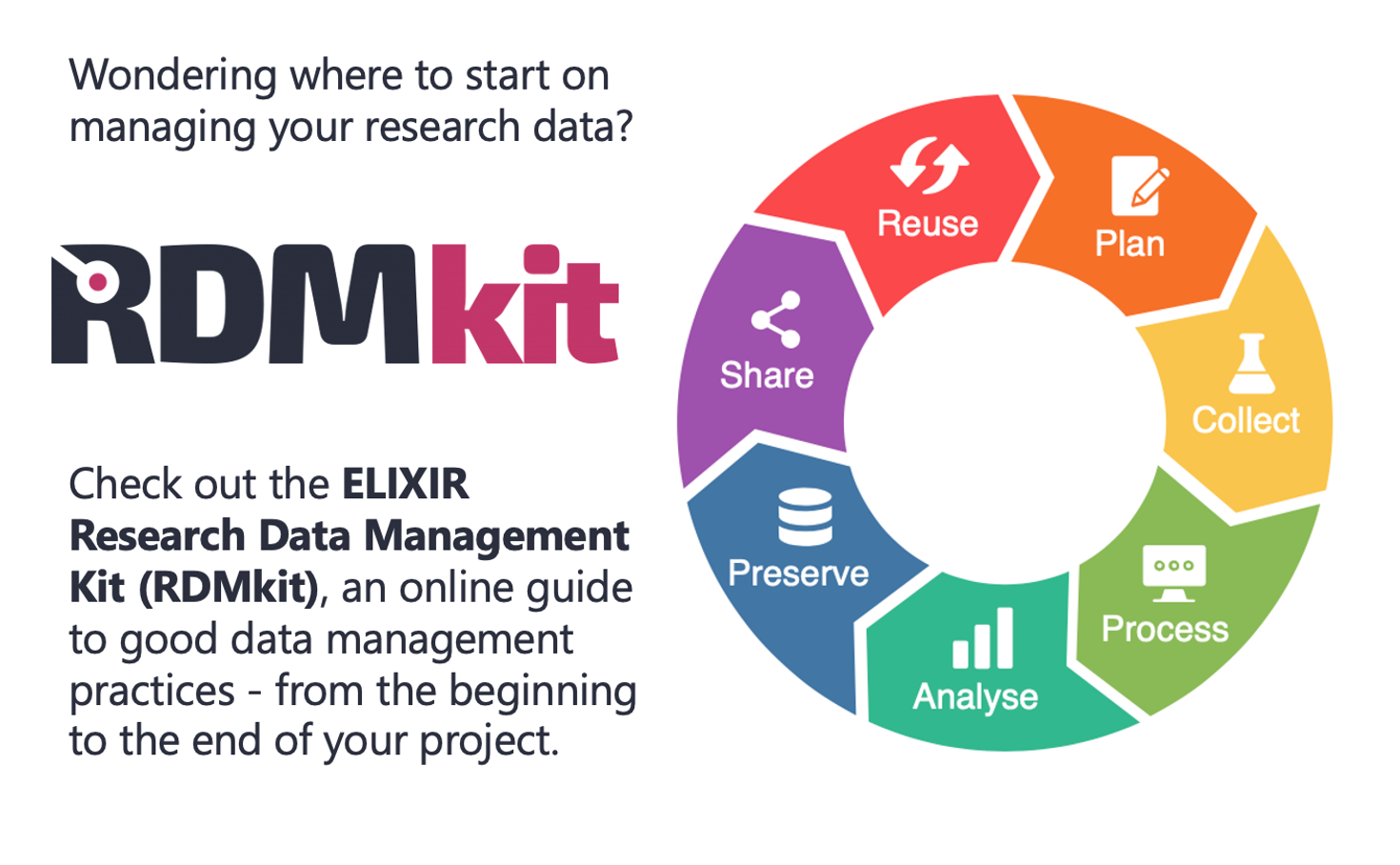
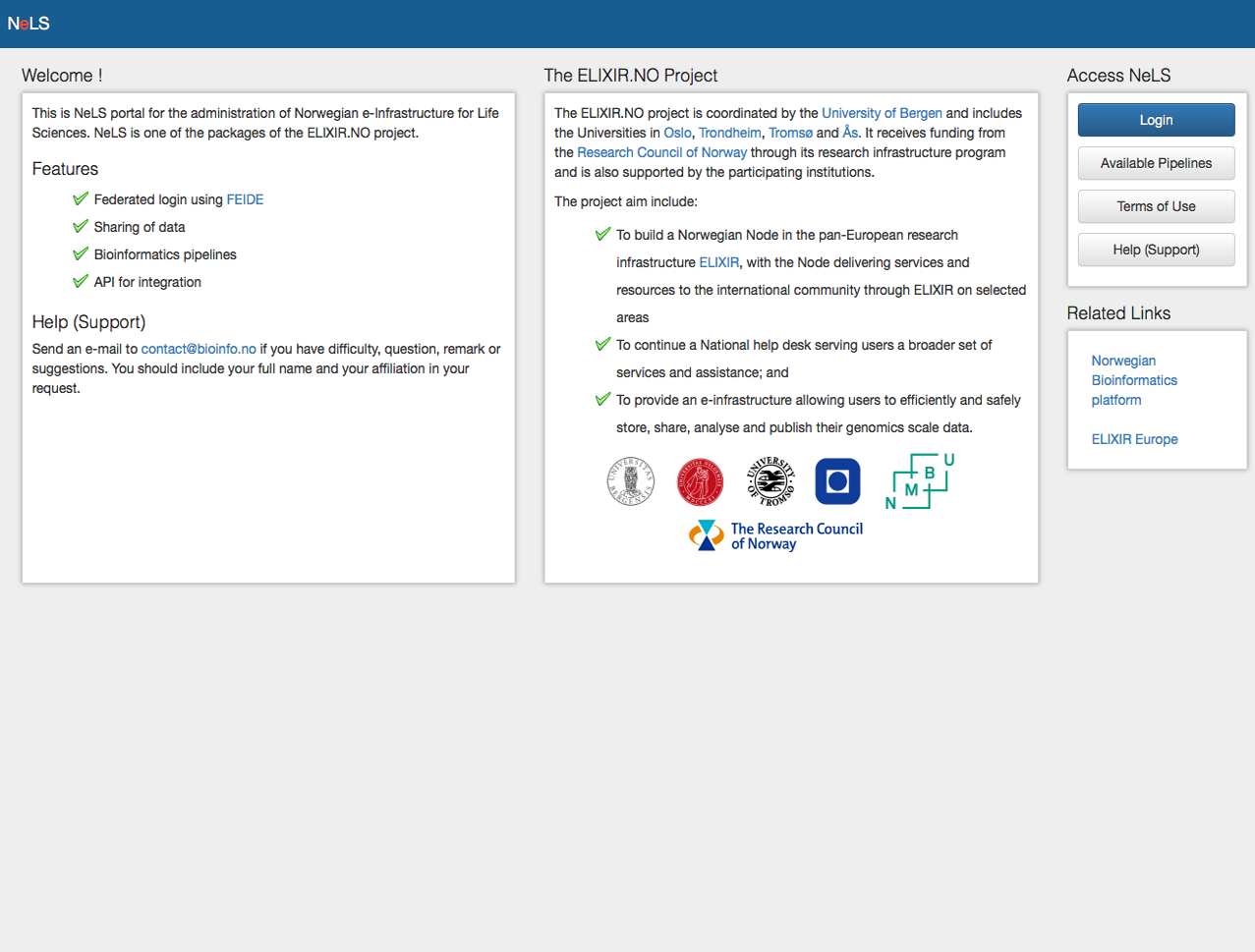
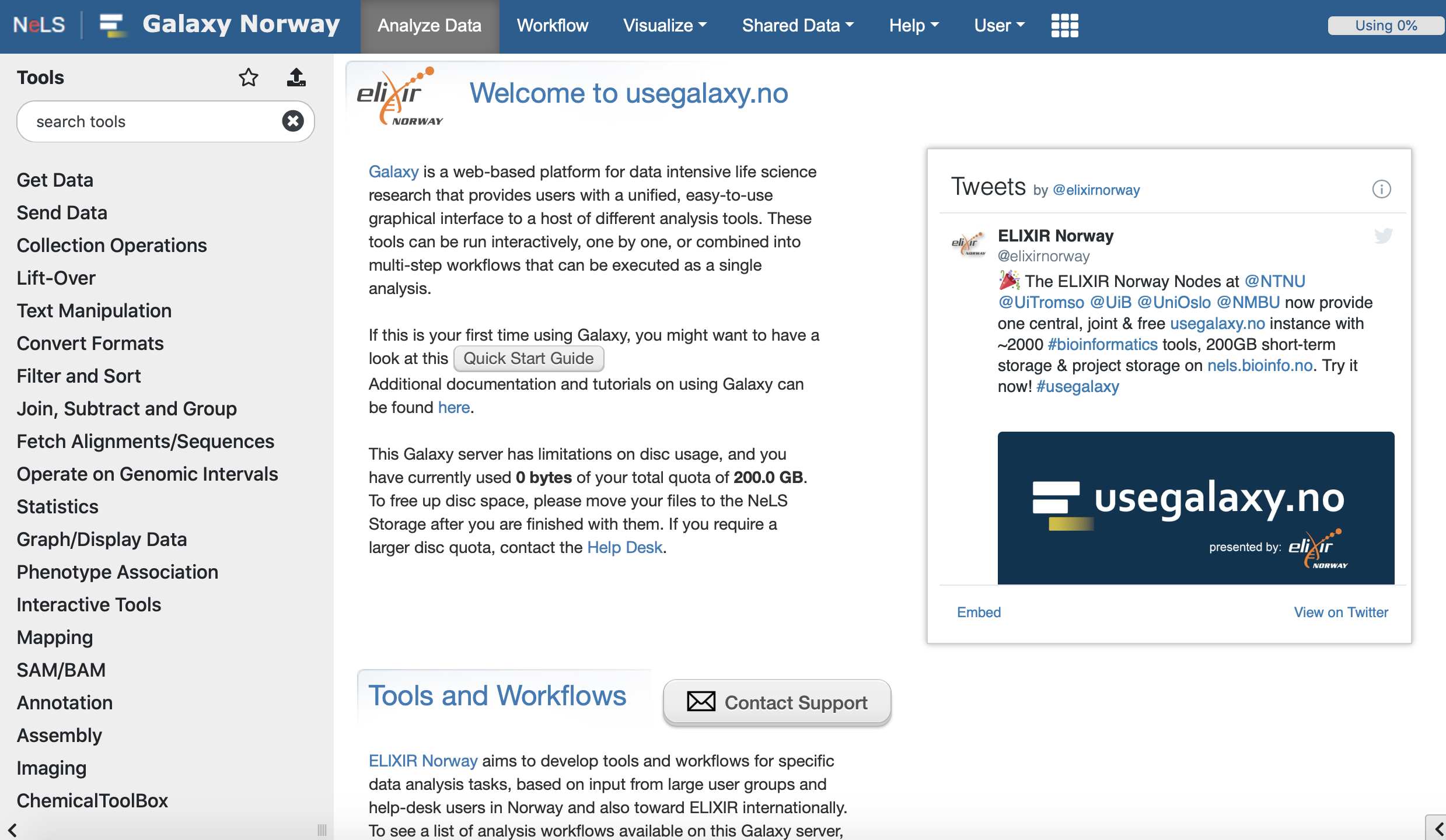
NeLS e-infrastructure
NeLS, the Norwegian e-Infrastructure for Life Sciences, for analysis, sharing and storage of high throughput genomics data.
Training
Local and national training in bioinformatics analysis and use of our tools.
International deliverables
ELIXIR Norway currently contributes and develops four services to ELIXIR Europe and the European community:
National nodes
ELIXIR Norway is a collaboration between five universities across Norway:
Research & Impact
- Publications
- Our members are involved in a wide range of high impact research projects and centres of excellence
- We collaborate closely with major research infrastructures and core facilities within biological and medical sciences in Norway


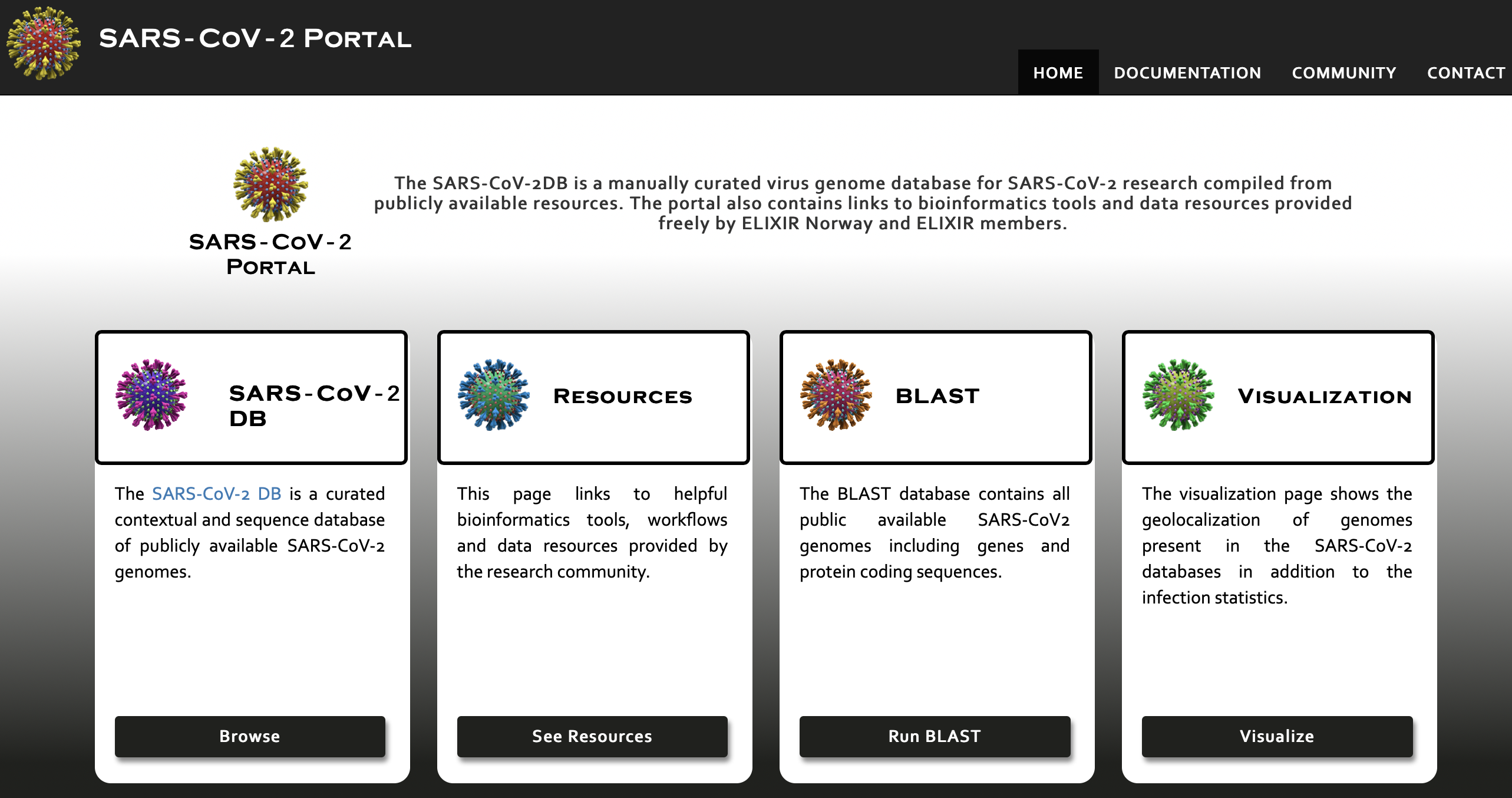
.png)